AI models for healthcare are proliferating, but most never leave the labs where they were designed. Real-world deployment is far more complicated than any multiple-choice graduate exam – hospitals use different systems, data formats, and security protocols that resist standardization. Kaapana, an open-source platform developed at the German Cancer Research Center (DKFZ), addresses these translation barriers by providing standardized infrastructure for medical AI research at scale. Currently powering AI workflows across German university clinics and major European cancer centers, the platform enables federated learning, where models train on distributed datasets without centralizing sensitive patient data.
We spoke with Kapaana’s lead engineers Ünal Akünal and Philipp Schader about the technical architecture, coordination challenges, and the surprisingly human reality.
Recently, there’s been a lot of talk about developing more AI models for healthcare. There are thousands of promising algorithms that support various clinical tasks, from triage to complex diagnostic cases. Most are trained on clean datasets and work beautifully in controlled lab environments, yet they rarely make it into the hospitals where they could actually be used. Most healthcare facilities lack the technical foundation to safely integrate external AI tools into their research workflows without months of custom engineering.
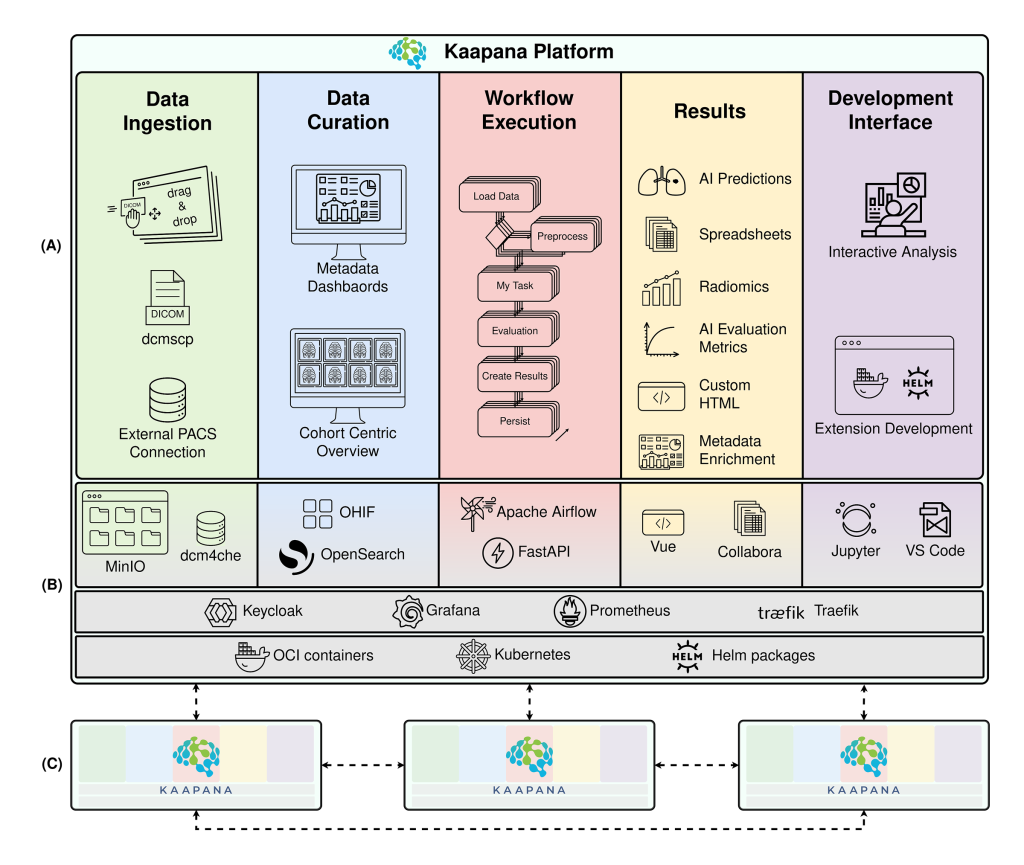
Enter Kaapana, an open-source platform developed at the German Cancer Research Center (DKFZ) in Heidelberg. It’s a tool for medical AI imaging research, contributing to collaborative infrastructure development at scale. It powers major centers across Germany and Europe.
We spoke with Ünal Akünal and Philipp Schader, lead engineers of Kaapana at the German Cancer Research Center, about how the platform operates and scales.
2Digital: For those of our readers who are not familiar with the concept, can you tell us a little bit more about Kaapana?
Ünal: Kaapana is an open-source platform for medical AI research, and our core mission is to fix the problem of translating AI into clinics. That’s the highest-level problem we’re approaching, and we solve it on the infrastructure level.

Healthcare facilities often don’t have the standardized technical foundation to deploy advanced AI algorithms safely. Different sites use different tools, systems, and data formats. On top of that, there are multiple roles and users: radiologists want to curate data, developers want to create their algorithms directly on real-world data – all players have specific expectations. And organizations are having a hard time building unified, interoperable infrastructure where medical AI can actually function at scale.
Kaapana provides the infrastructure layer that enables researchers to deploy AI tools directly inside clinics without requiring months of custom engineering.
Everything is decentralized, open-source, and cloud-native. It’s built on industry standard technologies that developers are already familiar with, and it provides established ways to access and share data, share methods, and run pipelines – all in a structured, elastic manner that adapts to different medical AI use cases.
Think of it as a battery-included platform. When you deploy Kaapana, you get:
– Large-scale data curation capabilities built in;
– State-of-the-art methods for image segmentation and other AI tasks;
– Integrated research toolkits like JupyterLab and MITK, Slicer;
– Standardized infrastructure that accommodates radiologists, data scientists, and developers simultaneously.
Kaapana enables multicenter research studies. Developing generalizable AI requires access to diverse datasets across multiple institutions. In medical AI, data is inherently valuable, but so is diversity. Rare medical conditions often require researchers to aggregate data across many sites. Some patients and pathologies are simply too uncommon in any single institution.
Kaapana leverages real-world imaging data directly within the platform and connects multiple clinical sites to enable federated learning – one of the hottest topics in medical AI research today. But federated learning only one for the features that Kaapana provides. The platform also provides data curation tools, pipeline execution capabilities, and all the infrastructure you’d expect from a standardized, enterprise-grade system.
2Digital: Could you explain how federated learning is different from traditional centralized machine learning?
Philipp: With federated learning, you don’t need to centralize sensitive data at all. Instead, the learning process happens locally at each site. Each institution trains the model on its own data, which produces a model update. Later, only the model update is shared, while the raw patient data remain proprietary. No identifying information leaves the hospital.

These model updates from all participating sites are then aggregated at a central hub, where they’re merged. The combined model is then distributed back to all sites, and the process repeats over multiple rounds. There’s just no centralized repository where all data can be leveraged for training.
It’s no secret that vast amounts of medical data is scattered across and locked in different institutions. By connecting multiple sites in a federated approach, you deliver the model to the data, not vice versa, and collect the diversity of information necessary to build more generalizable models than you’d get from training locally on a single institution’s set.
Federated learning is just one of several powerful use cases of Kaapana. Other collaborative approaches are also available – it depends on your needs.
A decentralized approach isn’t without tradeoffs. If you could centralize all data, this would theoretically give you optimal performance, and federated learning can only match that ceiling, but never exceed it. However, in the real world, several challenges come into play:
– Technical infrastructure complexity. Different sites have heterogeneous IT infrastructure. You need to coordinate with administrators at each institution to open network ports, establish secure communication, and ensure systems can actually talk to each other. This coordination overhead is significant.
– Data quality variation. Sites use different imaging protocols and data curation standards. Before you can run a federated learning workflow, you need harmonization steps to ensure all data meets consistent quality standards.
– Methodological challenges. Federated learning methodology is still an active research area. There are multiple approaches to aggregating models and orchestrating the learning process itself. You need infrastructure flexible enough to experiment with different training scenarios and see what yields the best performance for your particular use case.
One of the strongest arguments for federated learning is that most data protection officers consider aggregated model weights as anonymized data. This means many of the restrictions that apply to sharing raw patient data just don’t apply here, at least currently.
2Digital: Today, one can argue that we are amidst a healthdata goldrush: everybody is hunting for the data, and this project seems like something completely opposite – well, we don’t need your data, we’re just facilitating learning. Apart from technical issues maybe there’s some else, like business or political barriers?
Ünal: From a business perspective, federated learning actually makes perfect sense because you bring the algorithm to the data, not the other way round. You eliminate the need for expensive data-sharing contracts and complex legal structures.
The technical setup is actually harder. In centralized ML, you grab all the data and run it yourself. With federated learning, you need to understand each institution’s infrastructure, data homogeneity, and annotation strategies. If they differ (they usually do) you need to harmonize and standardize them first. You have to really get in touch with people onsite – this can take months of conversations with data managers, IT administrators, and clinicians at each hospital.
The most important work happens before any model training begins. Everyone focuses on the federated learning algorithm itself, but the pre-learning steps are actually far more important. You need to understand local data practices, infrastructure constraints, and even their vacation schedules because if their IT team is away, they might not be able to participate.
Coordination overhead is substantial, yet despite these challenges, federated learning is an enabler for situations where learning would otherwise not be possible due to data privacy limitations. To handle these pre-learning complexities, Kaapana cares about infrastructure, including standardized data validation, protocol harmonization, and workflow orchestration, letting institutions automate their participation.
2Digital: It’s funny and reassuring when you say that, in the age of AI, the most important part is talking to people.
Ünal: Yes, this is one of the most difficult parts, and this is not going away for sure. You can automate, as much as you want,, but you need to be able to collaborate with people for your research.
2Digital: Can you go into concrete details and elaborate on where Kaapana has proven success?
Philipp: Kaapana has been proven useful mostly within the context of large research consortiums but also for individual public institutions, private companies, and even smaller research projects of medical AI researchers. We see value across several areas, and here are several of the most ambitious projects.
The RAdiological COOperative Network (RACOON) demonstrates how Kaapana works at a national scale. The project was launched during the pandemic to unite Germany’s university clinics against COVID-19, and now, the scope has widened. The initiative installed nodes in every participating hospital. With the help of Kaapana as one of the tools in the infrastructure. Much work was also done by our partners in terms of getting the hardware and network setup. As a result a decentralized network was established where standardized AI workflows can be applied to radiological data across sites.
RACOON became our proving ground for federated learning at scale. Hospitals could share AI methods. Beyond training models, we built monitoring capabilities – aggregating patient numbers, case counts, and quality metrics across the entire network.
Cancer Core Europe – Data Rich Clinical Trials (CCE_DART) – a peer-to-peer model. Seven comprehensive cancer centers across Europe got together to improve oncological trials, including the development of new imaging biomarkers for cancer research. The design of this consortium allowed centers to act as peers, sharing methods directly with each other.
Kaapana provided the infrastructure layer that let each center contribute methods and benefit from others’ innovations simultaneously. It’s more distributed, less hierarchical.
UNCAN-CONNECT – while the original UNCAN initiative defined the blueprint, UNCAN-CONNECT is a flagship European project building infrastructure accelerating cancer research across nations. It operates through a federated model of national nodes and a central hub, and we are excited that Kaapana is embedded in the ecosystem.
2Digital: What infrastructure is needed to support Kaapana? Would adopting it require hiring specialized staff?
Ünal: It really depends on what you want to do. If you’re running a long-term project – say, something that’s supposed to operate for several years, rather than a single study – then yes, you need dedicated IT personnel. You’ll want a team that can take full ownership of your Kaapana instance: maintain it, perform updates, manage storage, and ensure everything runs reliably over time. The scale of your data also matters. If you’re handling terabytes of imaging data, someone needs to perform regular maintenance.
But there are other use cases. Many of our users are individual researchers or PhD students who want to build an end-to-end research pipeline on their own – without involving institutional IT. Here you can simply deploy Kaapana locally on your laptop, process your data, run experiments, and then remove the environment when you’re done.
It can be that lightweight and flexible.
Over the past five years, we’ve proven that Kaapana can provide value across that entire spectrum – from large-scale enterprise installations to small, independent research setups.
2Digital: There are multiple roles involved, but let’s focus on radiologists and IT administrators as examples. How long does it typically take them to become independently productive with this technology? What’s the realistic timeline?
Philipp: It depends on the role. For radiologists, we’ve built a streamlined web interface that’s quite intuitive. Think of it like the difference between Paint and Photoshop – if you’re unfamiliar with Photoshop, there’s a learning curve. But feedback from tech-savvy radiologists has been positive. They find the UI accessible and can quickly understand how to perform standard procedures like selecting datasets or sending outputs back to their workstations. There’s little to none ramp-up time if the interface is tailored properly to the use case.
For IT administrators, the timeline varies depending on their experience with cloud-native infrastructure. Those already familiar with Kubernetes, containerization, and modern DevOps tooling can debug issues and deploy Kaapana quite efficiently. Networking is often the biggest challenge – every institution has different network configurations and security policies. When problems arise, administrators who already understand how Kubernetes clusters handle networking can resolve them much faster.
2Digital: With the breakneck pace of technological development, we can see that even in Germany – a technologically advanced country – there’s now significant variability in technological preparedness. Ten years ago, hospitals were more or less on the same level.
Philipp: The environment has changed dramatically. For example, as security threats have become far more prominent, and hospitals now have to operate with much stricter protections. They work with security advisors who recommend specific technologies and configurations, which then get integrated into their networking infrastructure. In the end, we have a patchwork – one clinic adopts one solution, another uses something completely different, and they’re not necessarily compatible with each other.
Kaapana provides a common, harmonized layer that data scientists and researchers can rely on. All the entangled technicalities underneath – network configurations, communication protocols, security implementations – are abstracted away. The platform handles that complexity, so researchers can focus on developing and deploying AI methods.
2Digital: You are funded primarily by governmental research grants. In today’s situation, is it a strength or a weakness? What will happen if the funding diminishes?
Philipp: We’re predominantly funded by research grants because there’s a clear need for translational infrastructure in medical AI, and we address that need. In our experience, the trend is actually moving upward. More projects today are focused on translation, on getting AI from the lab into clinical practice. We’re glad we can help solve that problem, at least to some degree.
Even in multicenter studies without dedicated infrastructure you still need a budget allocated for data transfer and analysis. Our hope is that, by providing Kaapana and improving it with every project, we create cross-project value for the broader research community. If, at some point, there’s no longer funding because the need for translation infrastructure isn’t reflected in research priorities anymore, we hope the community we’ve built will help sustain the platform.
That said, we’ve also noticed the limitations of research grants. They’re time-bound, typically a few years, while infrastructure needs to operate on much longer timelines. There’s always the question: what happens when the grant ends? How do you extend the life of the infrastructure you’ve built – maybe by securing funding in the next phase of a grant? That’s an ongoing challenge in this type of financing model.


